File System
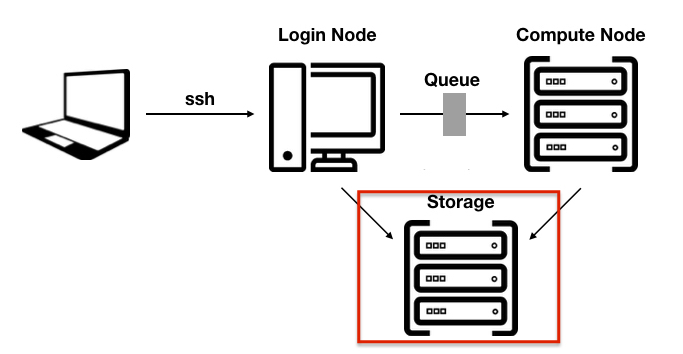
On a HPC cluster you have often several file systems. Different tasks need to be performed on different file systems.
Learning Objectives
- Get an overview about the different file systems on Euler.
- Know for which taks you can use which file system.
Overview about the different file systems on Euler.
There is your personal Euler home directory (/cluster/home/<USER>or ${HOME}), where you can keep scripts or own installed tools. Your personal scratch (/cluster/scratch/<USER> or ${SCRATCH}) is your working directory. Most of the bioinformatic tools need to read and write a lot of files e.g. I/O limited. The I/O is hgher on the SSD scratch drives, thus having the data (e.g. fastq or bam files) on the scratch often speeds up the analysis and reducing significantly memory usage. Data will be automatically deleted after two weeks. Thus important files have to be copied to GDC home or GDC projects for save-keeping. It is not allowed to directly output data to GDC home or GDC projects. Your GDC home is located here (/cluster/work/gdc/people/<USER>). You might also have access to one or more GDC project folders (/cluster/work/gdc/shared/<p999>). Be aware that these folders are shared with other users.
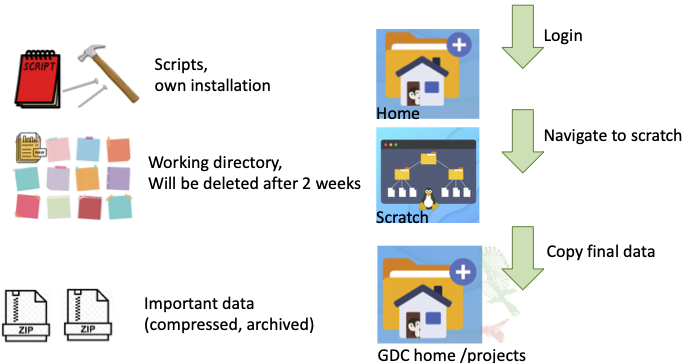
Personal Home

Variabe: ${HOME}
Keep scripts or own installed tools.
Personal Scratch - Your working directory

Variable: ${SCRATCH}
The fast SSD drive often speeds up analysis and reduces resource consumption. It is often useful to copy the files that you like to process to the scratch. Data is automatically deleted after two weeks.
Work file system (e.g. GDC home / GDC projects)

Here you can save your files that you like to keep.
- Keep it tidy and do not keep redundant data.
- Many small files (> 1,000) archived (taror zipped) to keep the number of files (inodes) low.
- There is a data storage fee at the end of the year.
- Euler should be not used as a data archive. After an analysis has been finished, we highly recommend to transfer your data to your long term storage (LTS) to preserve it in a long term perspective.
Challanges
You like to install a new tool. Where you install it?
We install the tool in the personal home.
You like to do read mapping and generate a lot of temporary files (sam, unsorted bam, final bam, fastq-files). The raw data is in GDC projects.
Copy the fastq files to your scratch. Do the read mapping on the scratch (sam, unsorted bam, unfiltered bam). We only copy the final filtered bam file to GDC projects.
We do some simulation in R and generate many small files. I like to keep them for a while.
Work on the scratch. Do the simulations. Archive the data (zip, tar) and copy the archive to your GDC home.
We like to to a SNP calling. The bam files are in your project folder
Copy the bam files to your scratch. Runn the tool on each chromosome. Copy the compressed raw vcf file to your project folder.