OSU-Mapping
Overview
- OTUs: In a fist step, I de-replicate the references to get unique sequences, which I call operational sequences units (OSUs).
- Mapping: Next, I map the cleaned reads to the OSUs using different identity (ID) and query-coverage (QC) thresholds.
BAFU 12S Reference Sequences ➜ OSUs
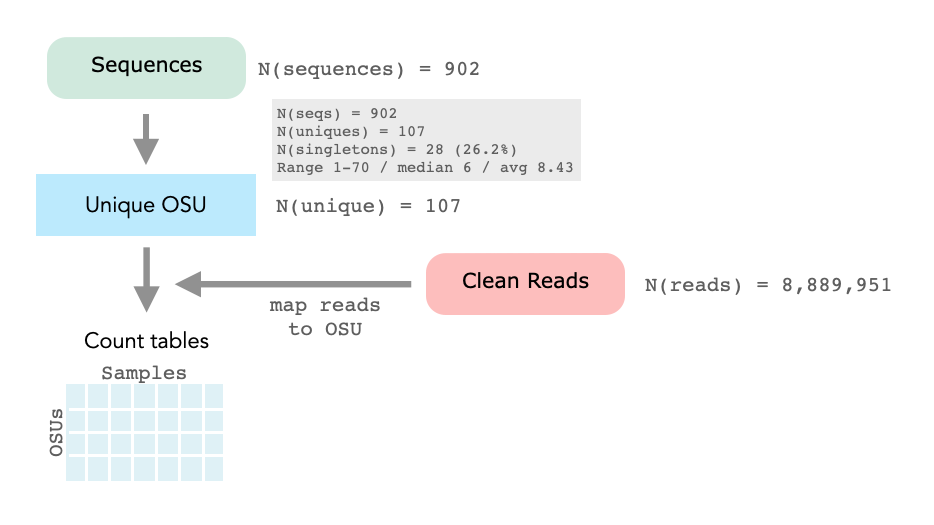
There are 902 fasta sequences in the BAFU 12S MiFish reference. After de-replication we have 107 (11.9%) unique sequences. We will call these unique refernces OSU (operational sequence untis) and for the mapping, we only use these unique sequences.
We have adjusted the sequence headers of the OSUs as follows. If all united sequences belong to the same species, the species name was taken over. The number of sequences that were merged was recorded with the note n at the end. In addition, the number of groupings per species was also noted.
Here an example Alburnoides_bipunctatus:
>Alburnoides_bipunctatus_1of2_n9
There are two (1of2) groups (clusters) of unique sequences for Alburnoides bipunctatus. The first cluster (1of2) contains 9 sequences.
Clusters with a mix of species such as Salmo were designated sp.
>Salmo_sp_1of3_n47
Mapping
For the read mapping we have two important parameters available: Sequence identity (ID) and query coverage (QC). We aim for highly accurate (1% ∼ 1 mismatch) and long (1% ∼ 2 nt) alignments. Both parameters will influence the number of reads that can be mapped to the OSUs. Stringent mapping criteria reduce mapping efficiency while relaxed ones will cause false assignment.
Special Cases
If there is a tie better two or more OSUs the read count goes to the first sequences (drip down). OSUs without counts are removed. Furthermore, mis-match positions are irrelevant. A potentially more informative nucleotide position is just another mis-match.
We use different values for identity (ID) and query coverages (QC) thresholds to find good mapping parameters.
| ID | QC | Reads Mapped | % Mapped | N(OSU) |
|---|---|---|---|---|
| 1.00 | 1.00 | 4,592,035 | 51.7 | 87 |
| 1.00 | 0.99 | 4,592,105 | 51.7 | 87 |
| 1.00 | .098 | 4,592,107 | 51.7 | 87 |
| 1.00 | 0.97 | 4,592,107 | 51.7 | 87 |
| 0.99 | 0.10 | 6,817,133 | 76.7 | 91 |
| 0.99 | 0.99 | 6,817,272 | 76.7 | 91 |
| 0.99 | 0.98 | 6,817,276 | 76.7 | 91 |
| 0.99 | 0.97 | 6,817,278 | 76.7 | 91 |
| 0.98 | 1.00 | 7,540,240 | 84.8 | 92 |
| 0.98 | 0.99 | 7,540,417 | 84.8 | 92 |
| 0.98 | 0.98 | 7,540,425 | 84.8 | 92 |
| 0.98 | 0.97 | 7,540,429 | 84.8 | 92 |
| 0.97 | 1.00 | 7,624,965 | 85.8 | 92 |
| 0.97 | 0.99 | 7,625,190 | 85.8 | 92 |
| 0.97 | 0.98 | 7,625,210 | 85.8 | 92 |
| 0.97 | 0.97 | 7,625,214 | 85.8 | 92 |
Identity (ID) threshold has a bigger influence on the mapping rate than query coverage (QC). The best results were obtained with the default values ID:97 and QC:100.
Here are the 15 OSUs to which no read could be mapped. With one exception (Alburnus arborella), these OSUs correspond to missing species in this dataset.:
- Alburnus_arborella_2of2_n1
- Alosa_sp_1of1_n8
- Ameiurus_melas_1of1_n5
- Cobitis_bilineata_1of1_n18
- Lampetra_planeri_1of1_n4
- Micropterus_salmoides_1of1_n5
- Neogobius_kessleri_1of1_n2
- Padogobius_bonelli_1of1_n9
- Rhodeus_amarus_1of3_n3
- Rhodeus_amarus_2of3_n11
- Rhodeus_amarus_3of3_n1
- Rutilus_pigus_1of2_n1
- Rutilus_pigus_2of2_n9
- Sabanejewia_larvata_1of1_n1
- Silurus_glanis_1of1_n12
OSU Extension
About 15% of the reads cannot be mapped to the OSUs. There are several reasons that could explain this observation. One possible reason could be the lack of species in the reference. For example, there are positive controls with cod in the dataset that are missing in the reference. For this reason, we extend our OSUs with consensus sequences of Gadus chalcogrammus and Gadus morhua. We also know from the OTUs annotation that we have additional sequences that are not from fish. For this reason, we also add some additional potential target sequences to the OSU mapping reference.
- Homo sapiens
- Sus scrofa
- Sturnus vulgaris
- Rupicapra rupicapra
- Ichthyosaura alpestris
- Bos taurus
- Turdus_merula
With these additional species we expand the OSUs reference to 116 sequences.
| ID | QC | Reads Mapped | % Mapped | N(OSU) |
|---|---|---|---|---|
| 1.00 | 1.00 | 4,683,758 | 52.7 | 95 |
| 1.00 | 0.99 | 4,684,009 | 52.7 | 95 |
| 1.00 | 0.98 | 4,684,012 | 52.7 | 95 |
| 1.00 | 0.97 | 4,684,012 | 52.7 | 95 |
| 0.99 | 1.00 | 6,991,312 | 78.6 | 99 |
| 0.99 | 0.99 | 6,991,768 | 78.6 | 99 |
| 0.99 | 0.98 | 6,991,773 | 78.6 | 99 |
| 0.99 | 0.97 | 6,991,775 | 78.6 | 99 |
| 0.98 | 1.00 | 7,753,236 | 87.2 | 101 |
| 0.98 | 0.99 | 7,753,792 | 87.2 | 101 |
| 0.98 | 0.98 | 7,753,802 | 87.2 | 101 |
| 0.98 | 0.97 | 7,753,806 | 87.2 | 101 |
| 0.97 | 1.00 | 7,877,395 | 88.6 | 101 |
| 0.97 | 0.99 | 7,878,060 | 88.6 | 101 |
| 0.97 | 0.98 | 7,878,082 | 88.6 | 101 |
| 0.97 | 0.97 | 7,878,086 | 88.6 | 101 |
The mapping results changed, albeit only slightly. However, the number of OSUs with read counts increased for all settings and so did the mapping efficiency. The OSUs without counts (n=15) did not change as expected.
Mapping Analysis
Positive Control Samples (PCP)
The most dominate species in the positive samples (PCP) is Atlantic cod (Gadus morhua). We have to keep in mind that we only have two species of cod (Gadus morhua and Gadus chalcogrammus) in the reference. Although the positive sample contains only one species, we also find other as well.

Negative Control Samples (FCN and PCN)
There are only two (filter) negative samples with significant (>50) number of total counts. The composition of the two negative samples is different. The Lake trout (Salvelinus namaycush) is unique to sample FCN-1A but the ZOTU with MitoFish-Plus annotation shows bacterial contamination of about 10%. Negative sample FCN-1C contain Squalius cephalus and Salmo carpio to equal parts. Again, ZOTU with MitoFish-Plus annotation shows bacterial contaminant.

>ZOTU78 (Alpha proteobacterium - Roseomonas?)
AGCCGCGGTAATACGAAGGGAGCTAGCGTTGCTCGGAATGACTGGGCGTAAAGGGCGCGTAGGCGGCGGCGTAAGTCAGA
TGTGAAATTCCTGGGCTTAACCTGGGGGCTGCATTTGAGACTGCGTTGCTAGAGGACGGAAGAGGCTCGTGGAATTCCCA
GTGTAGAGGTGAAATTCGTAGATATTGGGAAGAACACCGGTGGCGAAGGCGGCGAGCTGGTCCATTACTGACGCTGAGGC
GCGATAGCGTGGGGAG
>ZOTU81 (Sediminicoccus rosea - Acetobacteraceae?)
AGCCGCGGTAATACGAAGGGGGCTAGCGTTGCTCGGAATGACTGGGCGTAAAGGGCGCGTAGGCGGCACAACTCGTCAGG
CGTGAAATTCCTGGGCTTAACCTGGGGGCTGCGTTTGATACGGTTGAGCTAGAGGATGGAAGAGGCTCGTGGAATTCCCA
GTGTAGAGGTGAAATTCGTAGATATTGGGAAGAACACTGGTGGCGAAGGCGGCGAGCTGGTCCATTACTGACGCTGAGGC
GCGAAAGCGTGGGGAG
Samples
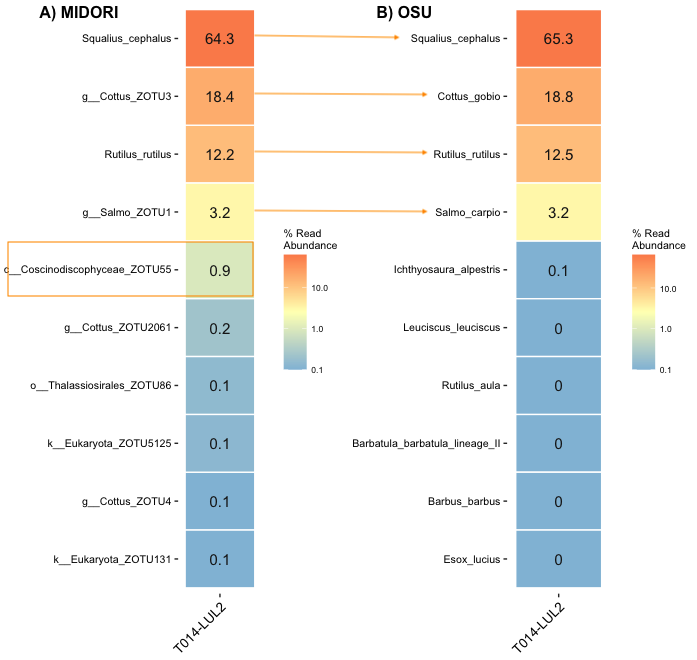
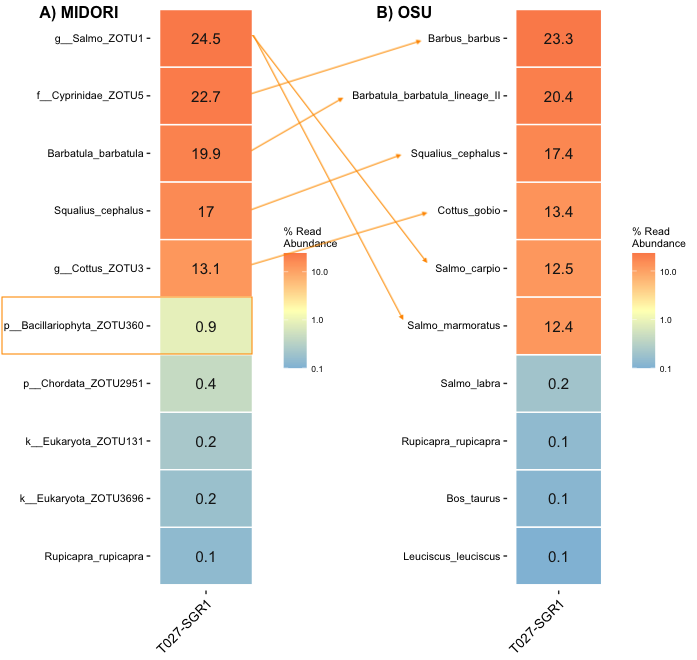
Pairwise sample comparisons show that the composition is similar for the ZOTUs and for the OSUs approach. Differences exist for the rare zOTUs and OSUs, respectively. OSUs always have species indications, but this could give a false sense of security. In addition, the reference restricts species recognition for the OSU approach, while the zOTU approach is not restricted and has the potential to discover more species (e.g. 1% diatoms).