More Simulations with R
There are some nice R packages for learning population genetics. We will focus on the HardyWeinberg and learnPopGen packages, both of which have help options and documentation (see below).
R Packages for PopGen
HardyWeinberg
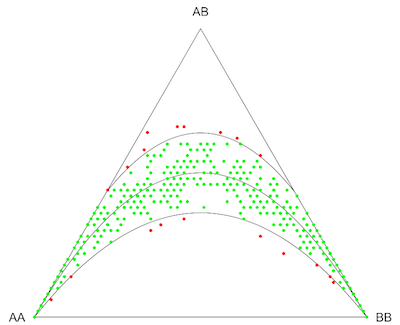
The HardyWeinberg package offers several functions for various tests (e.g. HWE), genetic marker data simulations, graphical tools (e.g. ternary plots) and provided data sets.
Example: Simulation for marker data
## Dimensions of the population(s)
Npop <- 100 # number of individuals
Nind <- 10 # number of popualtions
# Matrix with genotype counts:
pop <- HWData(Nind, Npop)
pop
learnPopGen
R package learnPopGen was developed to learn population genetics, quantitative genetics, and evolutionary theory.
Example: Migration, Selection, and Drift (msd)
learnPopGen::msd(p0 = c(0.5, 0.5), # starting frequency for the A1 allele
Ne = c(100, 100), # effective population
w = list(c(1, 1, 1), c(1, 1, 1)), # fitnesses of the three genotypes
m = c(0.05, 0.05), # rates of migration
ngen = 500, # number of generations
colors = c("red", "blue") # population color id
)
R Script with Examples
With the help of both packages, I have written some R code examples. It extends the do-it-yourself example by a lot. I wrapped everything into a Rmarkdown file. There should be no problem to run individual code chunks. I also knitted an HTML page in case you have difficulties to do it yourself. I hope you learn something from the code and constrictive feedback is appreciated.
I also provide a zip file containing the Rmarkdown script and the HTML file.
Note
The HTML report works well with the Internet browsers Brave, Firefox, etc. but there are problems with older versions of Safari. You can download the HTML file and open it locally.