Alpha Diversity Help¶
A quick summary to better understand alpha diveristy. To make this work, you need to download and import the dummy dataset described in chapter ”Phyloseq Help”.
## Subset Samples d.A <- subset_samples(d, GID == "A") d.B <- subset_samples(d, GID == "B") d.AB <- subset_samples(d, GID == "A" | GID == "B") d.C <- subset_samples(d, GID == "C") ## There might be OTUs without counts after subsetting data sum(taxa_sums(d.A) == 0) sum(taxa_sums(d.B) == 0) sum(taxa_sums(d.C) == 0) ## Remove the "zero" OTUs (if needed) # d.A <- prune_taxa(taxa_sums(d.A) > 0, d.A) ## OTU table overview t(otu_table(d.A))
| OTU1 | OTU2 | OTU3 | OTU4 | OTU5 | OTU6 | OTU7 | OTU8 | OTU9 | OTU10 | OTU11 | OTU12 | OTU13 | OTU14 | OTU15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SA1 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 |
| SA2 | 20 | 10 | 10 | 5 | 5 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| SA3 | 34 | 16 | 16 | 8 | 8 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| SA4 | 82 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| SA5 | 90 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
## Alpha-Diversity Table alpha.diversity <- estimate_richness(d.A, measures = c("Observed", "Chao1", "Shannon", "InvSimpson")) round(alpha.diversity, 3) ## A nice table #DT::datatable(round(alpha.diversity, 3))
| Observed | Chao1 | se.chao1 | Shannon | InvSimpson | |
|---|---|---|---|---|---|
| SA1 | 9 | 9 | 0.000 | 2.197 | 9.000 |
| SA2 | 9 | 15 | 7.123 | 1.729 | 4.459 |
| SA3 | 9 | 9 | 0.000 | 1.751 | 4.470 |
| SA4 | 9 | 37 | 21.221 | 0.485 | 1.203 |
| SA5 | 1 | 1 | 0.000 | 0.000 | 1.000 |
A table might be of limited use for larger data sets.
## Alpha-Diversity Plot plot_richness(d.A, measures = c("Observed", "Chao1", "Shannon", "InvSimpson")) ## A "nicer" ggplot based plot p <- plot_richness(d.A, color = "GID", measures = c("Observed", "Chao1", "Shannon", "InvSimpson")) p + geom_point(size = 5, alpha = 0.7)
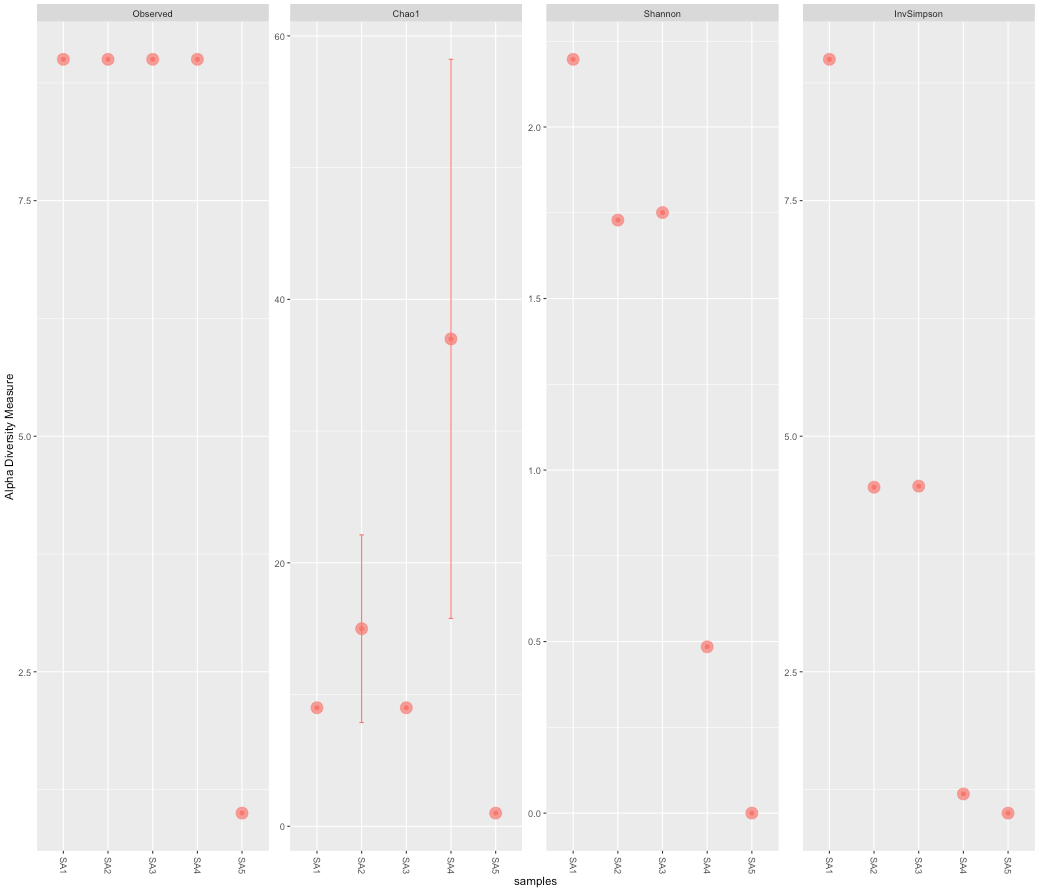
## Combined OTU and Diversity Tables cbind(t(otu_table(d.A)), alpha.diversity)
It is easy to change the counts and play around with different values.
## Change Counts otu_table(d.A)[,5] otu_table(d.A)[,5] <- c(seq(90,10,-10)) otu_table(d.A)[,5] otu_table(d.A) estimate_richness(d.A, measures = c("Observed", "Chao1", "Shannon", "InvSimpson"))