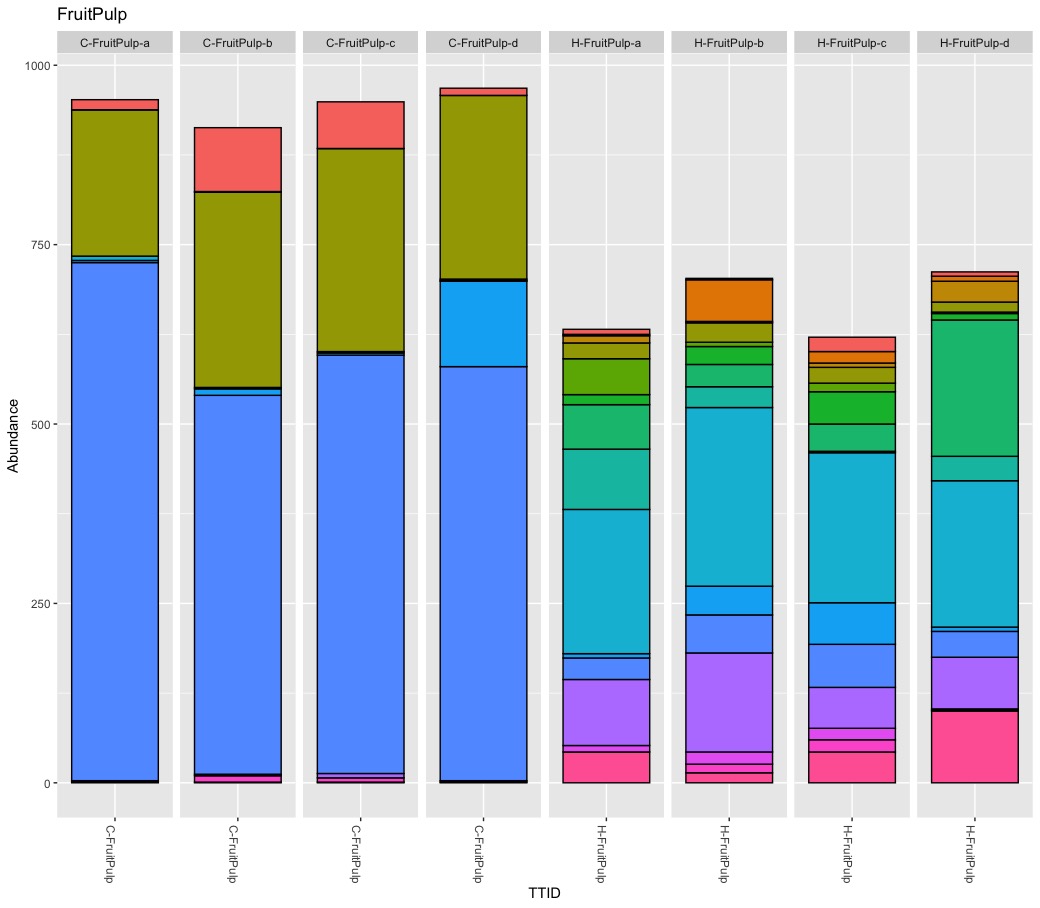
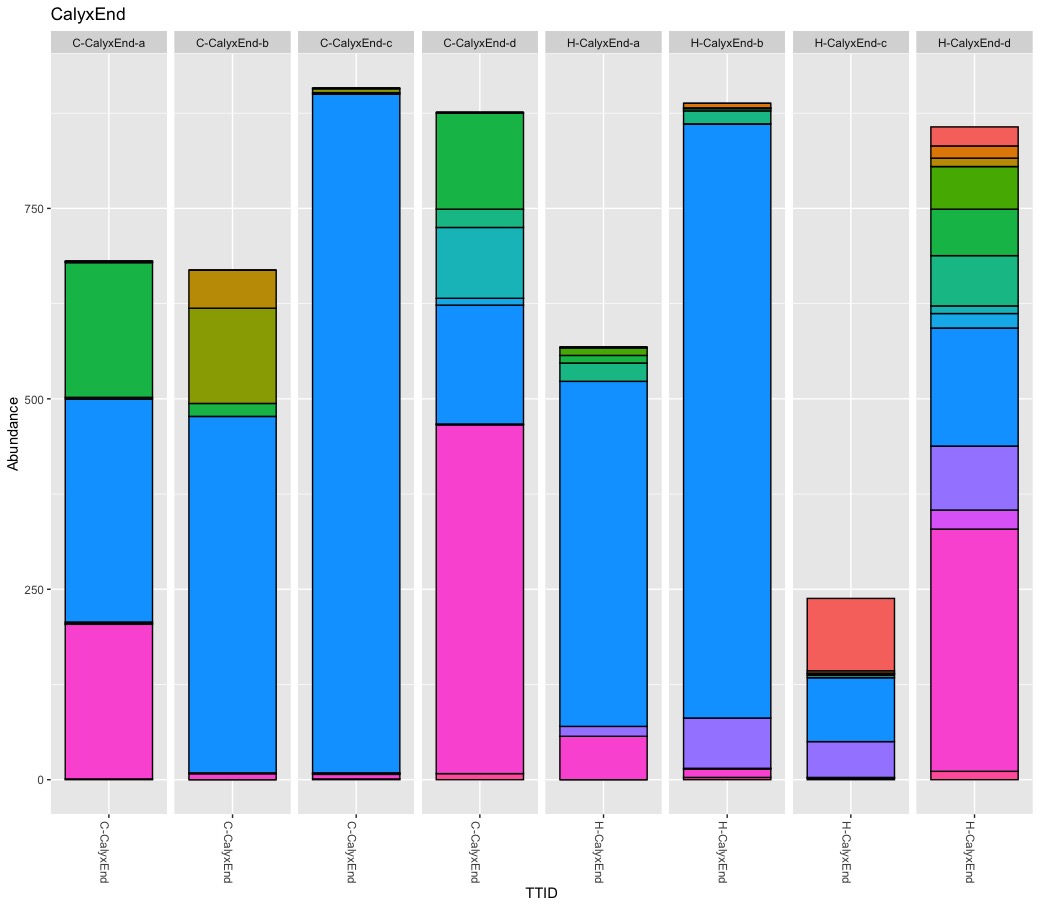
Microbiota Structure (bedtime story)
My re-analysis of the bacteria structure does not correspond with the authors’ results, I have problems following their conclusions, and I cannot support their claims. It seems that details are ignored to fit certain expectations.
Taxonomic composition¶
Burkholderiales¶
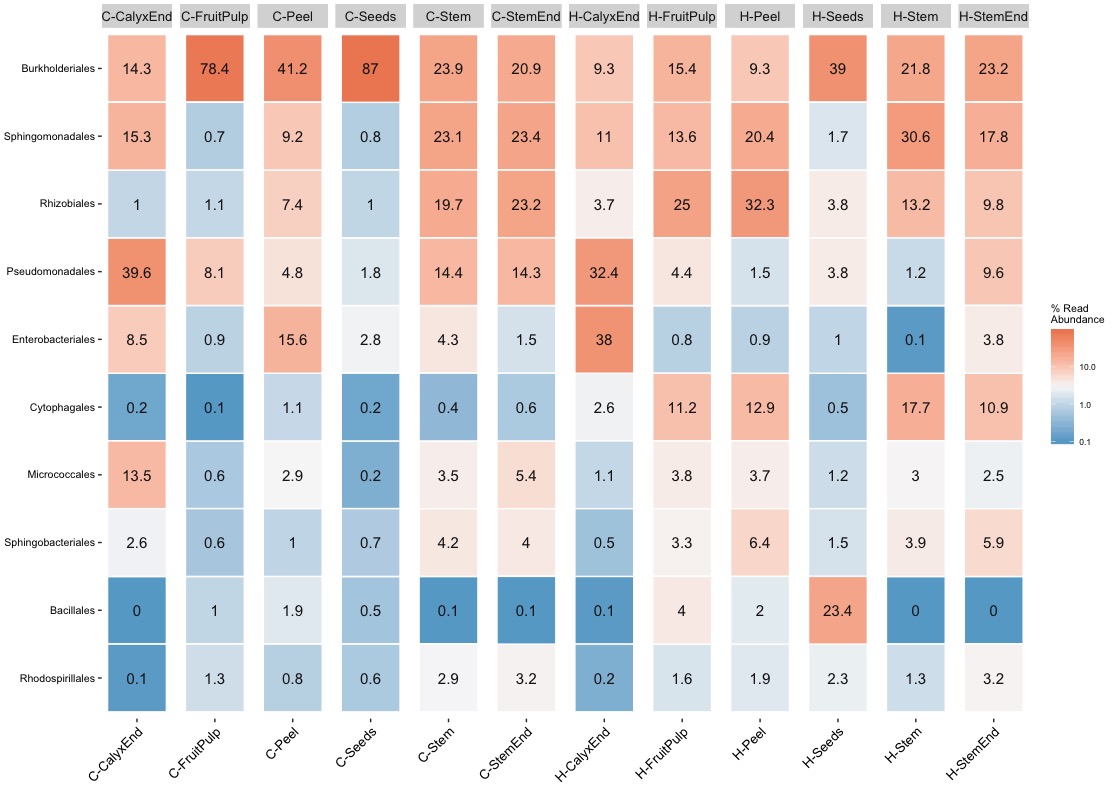
The discussion of the taxonomic composition is selective and therefore problematic. For example, the authors claim that Burkholderiales is dominant in conventional apples but a relevant detail is missing.
Conventional apple microbiota was furthermore found to be less even constructed and highly dominated by Burkholderiales, accounting to almost 43% abundance. source: Wassermann et al. 2019
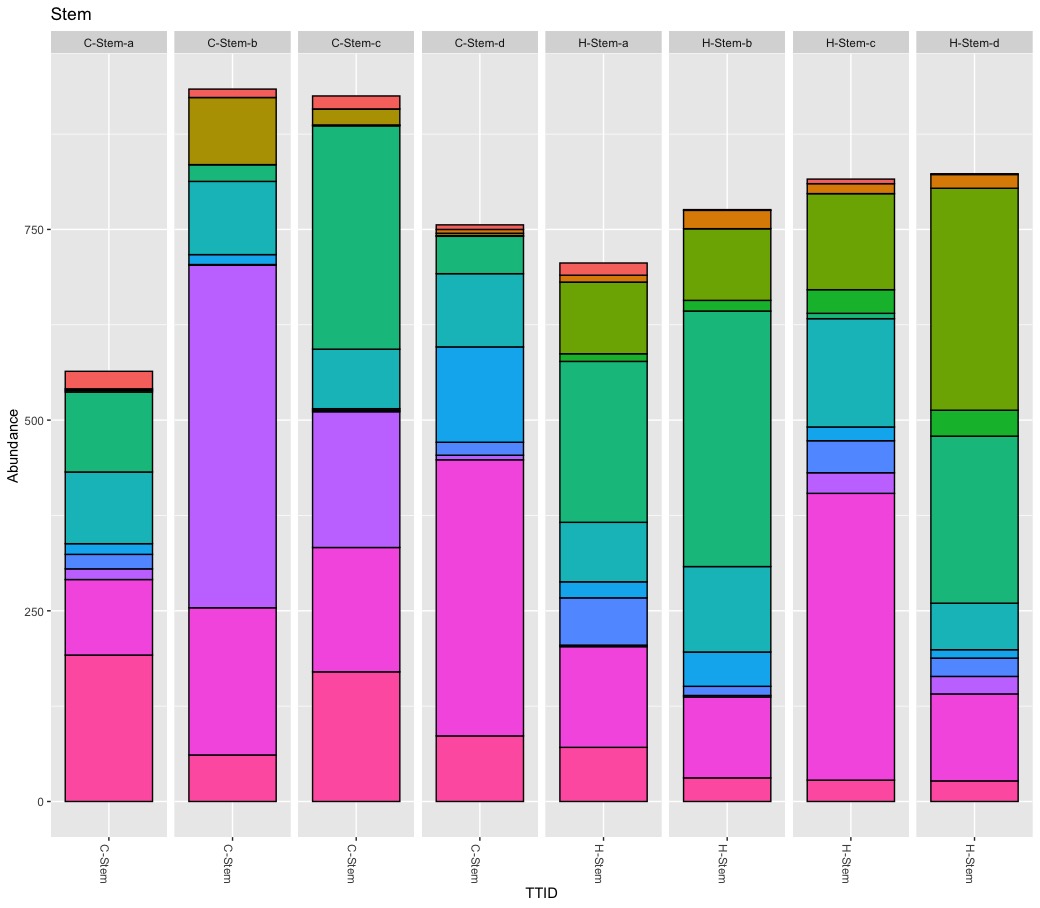
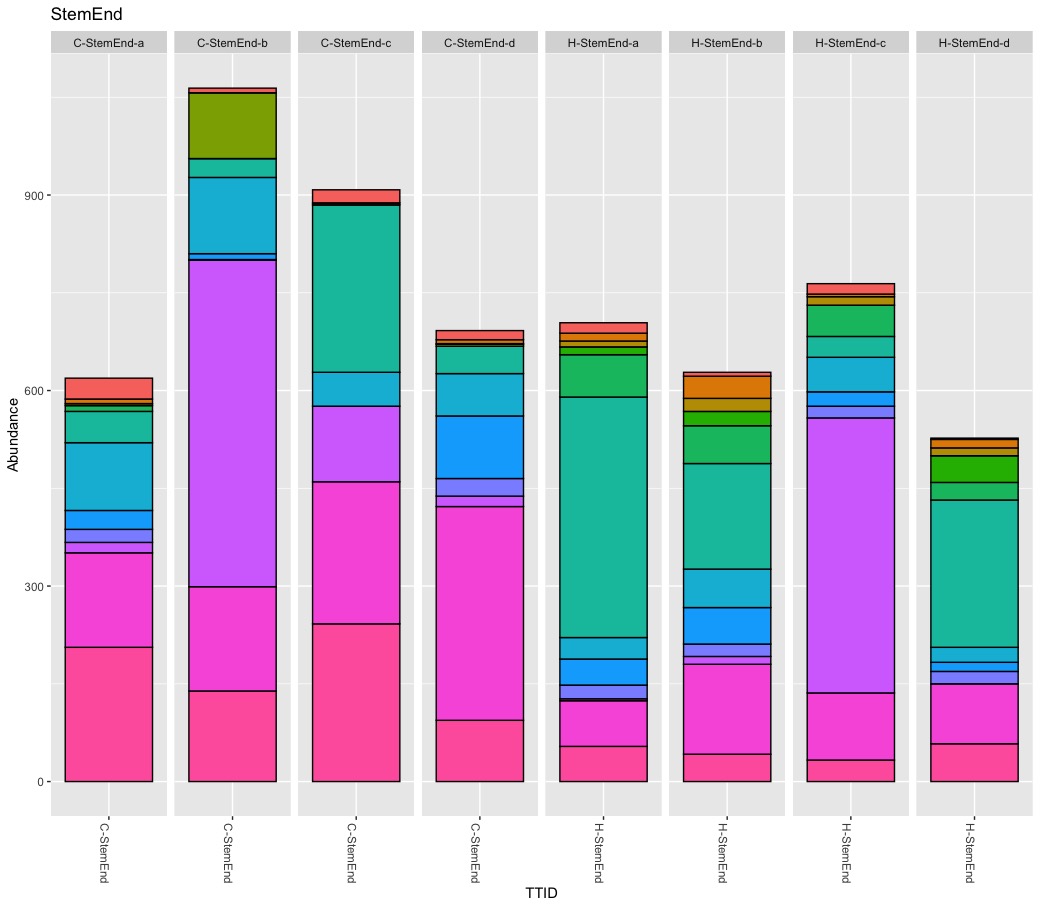
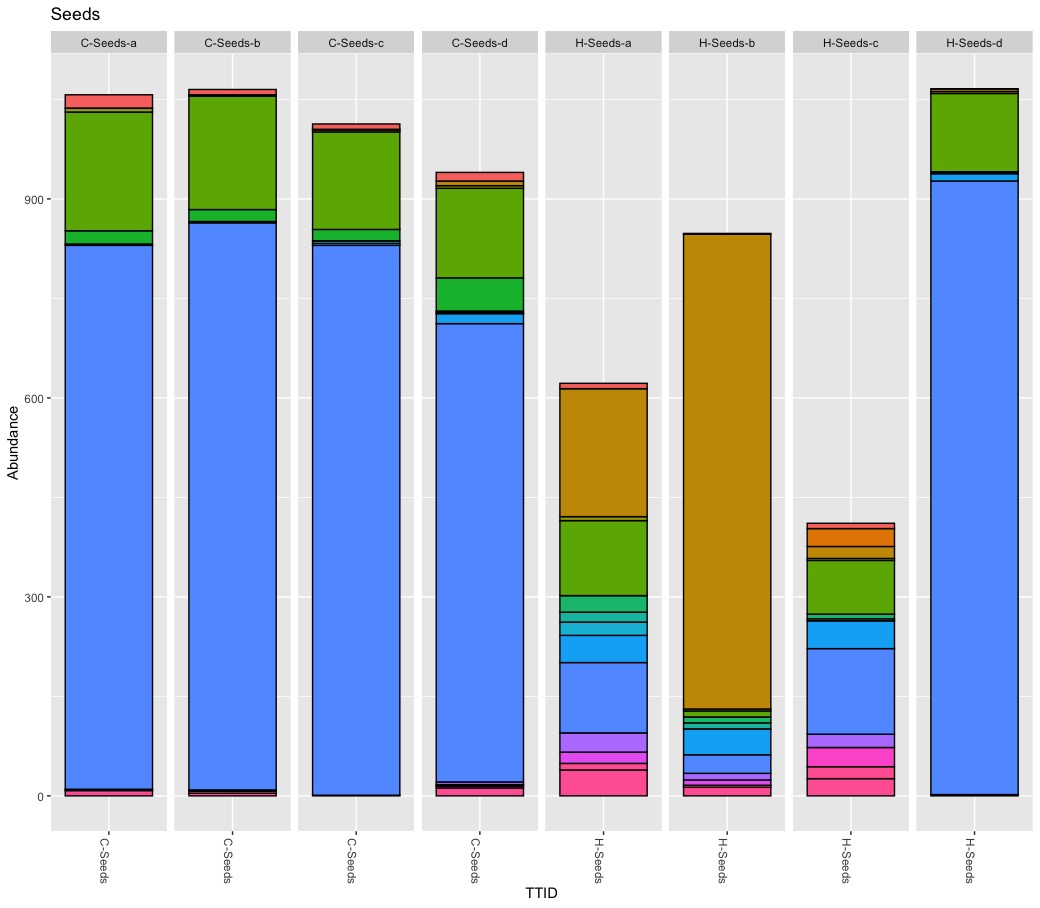
True, there are more Burkholderials in non-organic (45.5%) compare to organic (20.2%) samples but only because the bacteria order Burkholderials is dominant in the fruit pulp (80.1%) and the seeds (88.5%) of non-organic apples. There is no difference in stem or stem end.
Ralstonia¶
There is one OTU assigned to Ralstonia and it is abundant in fruit pulp, peel, and seed samples of supermarket apples. However, I found the highest abundance of 80% in one seed sample of an organic apple.
Enterobacteriales¶
I do not see why Enterobacteriales should be a signature taxa of conventional apples.
The order Enterobacteriales was one of the signature taxa of conventional apples as well; among them, we would like to highlight the almost ubiquitous occurrence of OTUs assigned to Escherichia-Shigella in the tissues of conventional apples (although low abundant) and their absence in organically managed apples. source: Wassermann et al. 2019
My results show similar levels of Enterobacteriales for non-organic (5.8%) and organic (7.5%) apples. Again, the pooled abundance does not reflect the variance between tissue. It turns out, Enterobarteriales is the most dominate bacteria order (38%) in calyx end samples independent of the treatment.
Erwinia¶
This is a truly rare find!
"Abundance of a not further assigned Enterobacteriaceae taxon (Enterobacteriaceae sp. in Figure 6), Erwinia and Escherichia-Shigella were significantly more abundant in conventional apples." source: Wassermann et al. 2019
First, Erwinia is very rare. In fact, it is absence in the defined “core” microbiota and I was only able to find it in one calyx sample of a supermarked apple.
Remark about the citation of "Pirhonen et al., 2018”. The study is focusing on Erwinia carotovora subsp. carotovora and it was published in 1993.
Escherichia-Shigella¶
What does it mean to have significantly more Escherichia-Shigella on conventional apples compared to organic apples?
“Escherichia-Shigella were significantly more abundant in conventional apples.” source: Wassermann et al. 2019
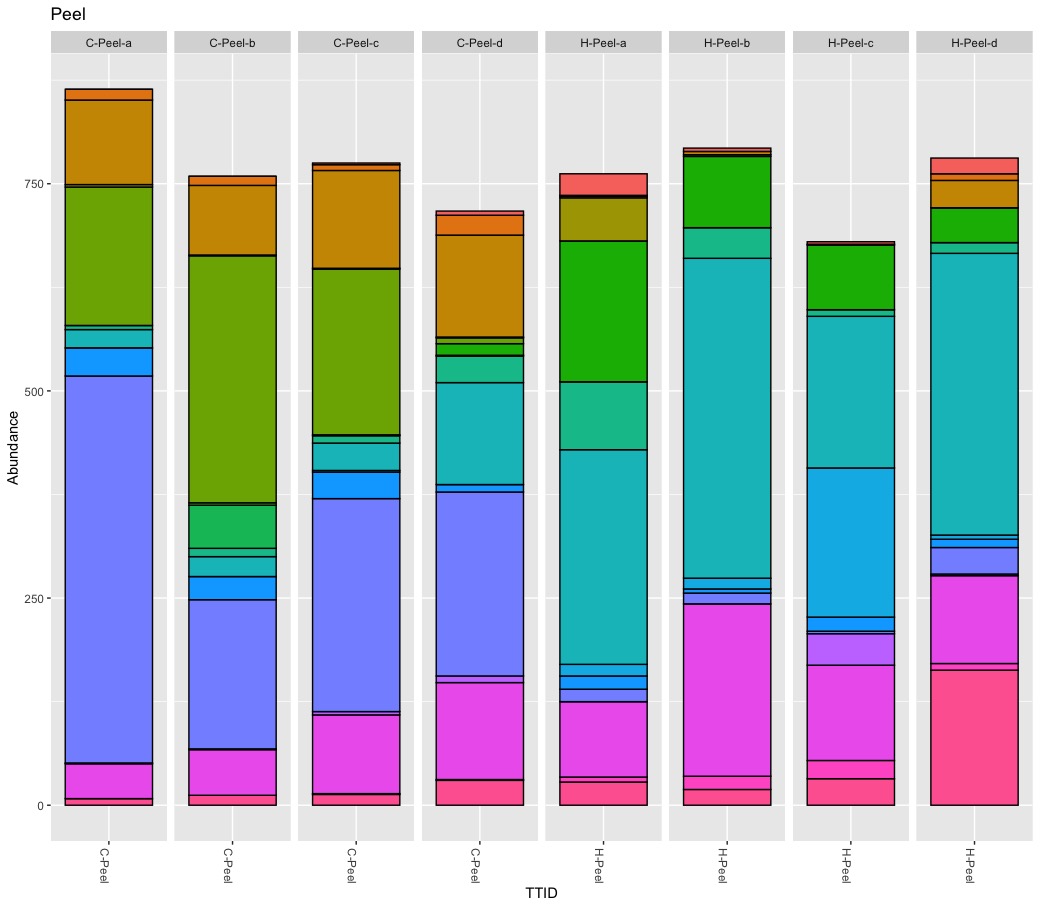
It is a tricky game to reduce abundance to present-absent for are relatively rare baterium like Escherichia-Shigella. Escherichia-Shigella is present in almost all samples expect stem of non-organic samples while it is absent in all samples from organic apples except for two fruit pulp samples. My findings seems to confirm the observed pattern in the paper but again the devil is in the detail. Important here is the abundance! The abundance for Escherichia-Shigella is low for all samples except for tissue peel of three replicates. I repeated the rarefying step and could only confirm stable results for the tissue of peel samples of non-organic samples.
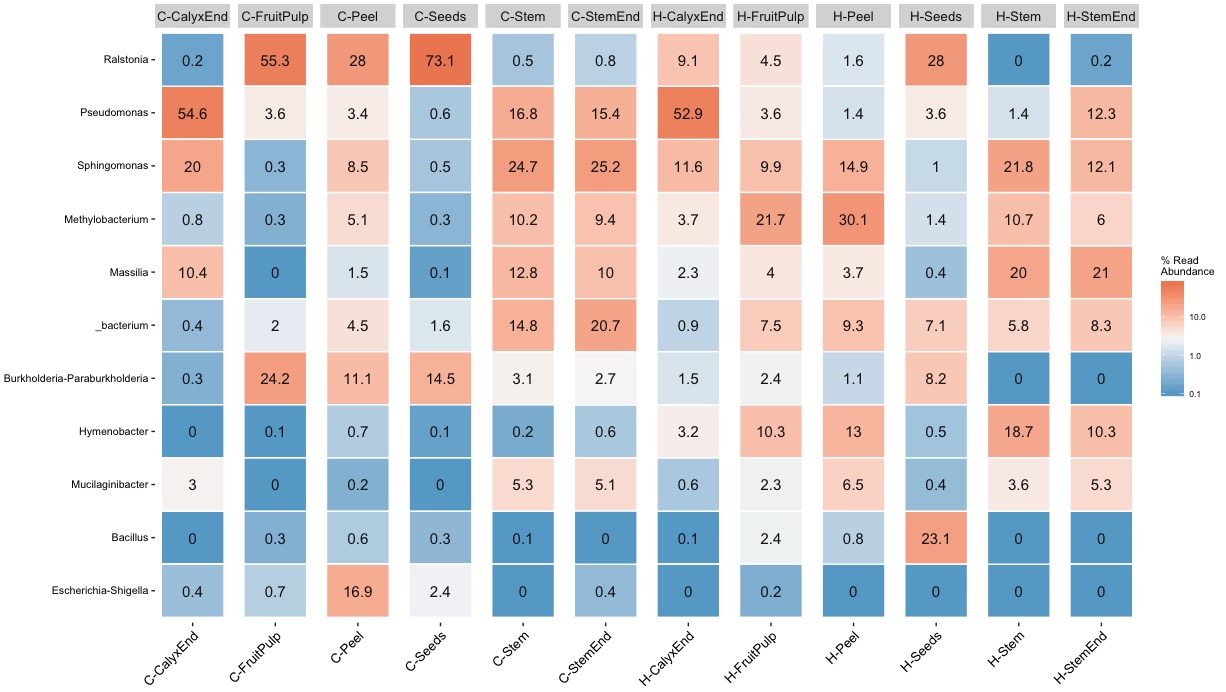
Lactobacillus¶
I feel uncomfortable with the “health claim” of organic apples as a source of probiotic bacteria.
“Controversially, Lactobacillus, which is frequently used within probiotics, was one of the core taxa of organic apples.” source: Wassermann et al. 2019
I cannot backup the claim about Lactobacillus. First, Lactobacillus is rare, very rare. The count ranges from 0 to 70 with a total abundance of 0.2%. Second, the highest abundance (6.6% “core” sample, 4.7% rarefied samples) was found in a single conventional peel sample. Because Lactobacillus is rare its presence-absence is affected during the rarefying step. Third, I never found Lactobacillus in all 4 biological replicates.
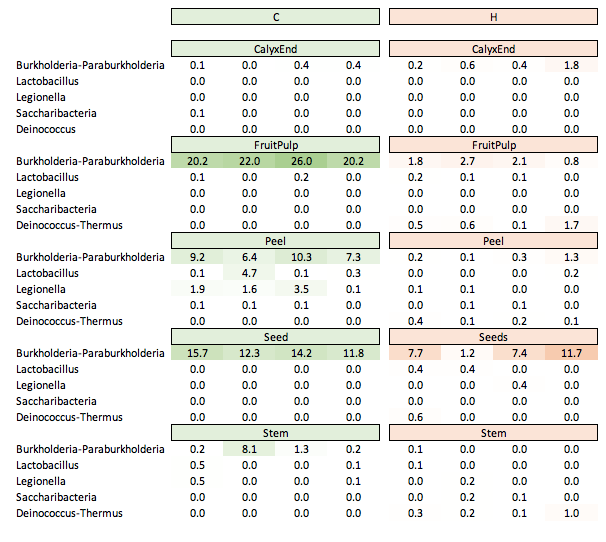
Legionella¶
The presence of Legionella got ignored by the authors. Escherichia-Shigella and Legionella show a very similar abundance pattern. Both occur at > 1.5% abudnace on the same three peel samples.
Burkholderia-Paraburkholderia¶
Another ignored but potentially interesting genus could be Burkholderia-Paraburkholderia
Summary¶
Rarefying samples (down sampling) is a random process, and as such it influence presence-absence and abundance of rare taxa. Although beautiful, Figure 5 is difficult to read and understand. Besides the too many colors, pie charts are nice for compositional data of one microbiome but not optimal to compare microbiomes (scaling). They author also pooled biological replicates, ignoring significant variation within replicates. I consider pooling samples in this study problematic. It seems the authors pooled biological replicates to increase diversity and pooled tissues to find differences between treatments. As previously shown, the within sample variation is substantial for some samples. I think it matters if (rare) taxa occur only once or are present in all replicates. I consider it wrong to define a core apple microbiome by an arbitrary abundance threshold (e.g., 0.01%) alone and to ignore prevalence at all. I also believe pooling tissues to compare treatments is risky because it leads to the wrong conclusions. It is interesting to know that potential harmful bacteria occur on non-organic apples but of limited use for health-related questions. Bacteria are not bad news and even species belonging to Escherichia-Shigella are mostly harmless. For practical consideration, I need to know if pathogens can be simply washed away or if they are part of the fruit itself.
Intra-Sample Variation for Top 30 OTUs¶